cerberus
Creating and visualizing more comprehensive transcriptome annotations
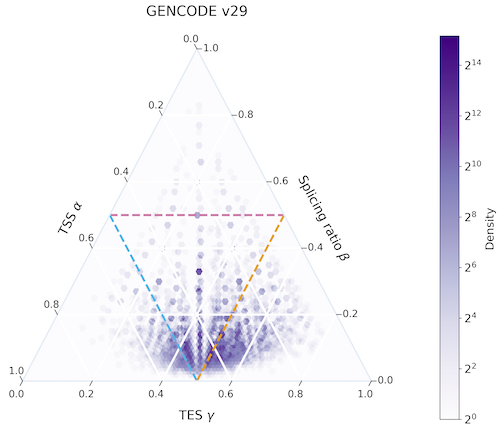
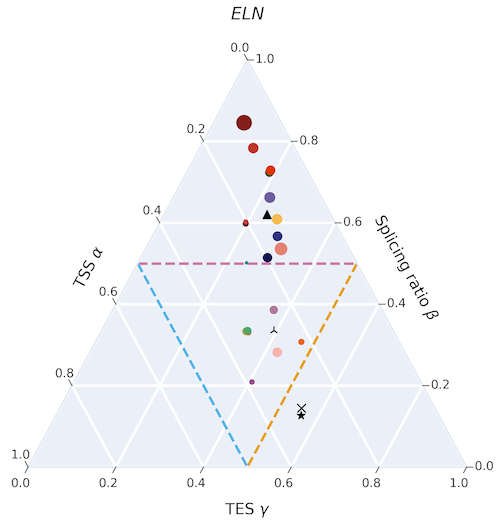
Cerberus is a set of tools used to integrate transcriptome information from a variety of input sources to create comprehensive transcriptome annotations. It can parse BED regions for 5’ and 3’ ends of transcripts and GTF files for unique annotated intron chains, and integrate them all into one collective source. This source can then be used to convert existing GTF transcriptomes to their cerberus version.
Cerberus transcriptomes are directly compatible with Swan, which can detect the annotated 5’ / 3’ ends and intron chains from cerberus and quantify expression of each of these features.
In the near future, tools to visualize transcriptional diversity based on each gene’s 5’ end, intron chain, and 3’ end diversity will be incorporated.